Class: BIOL 0440 – Inquiry in Plant Biology: Analysis of Plant Growth, Reproduction and Adaptive Responses
Instructor(s): Dr. Alison DeLong, Dr. Mark A Johnson
Student(s): Thomas Murphy, Paige Lind, Ryan Chaffee
Affiliated Faculty and Collaborators: Ann Loraine (UNC-Charlotte), Kelly Pan, Sorel Ouonkap Yimga
“We learned that genes could themselves be different while doing the same thing, and example of convergent evolution.”
-Student
Description:
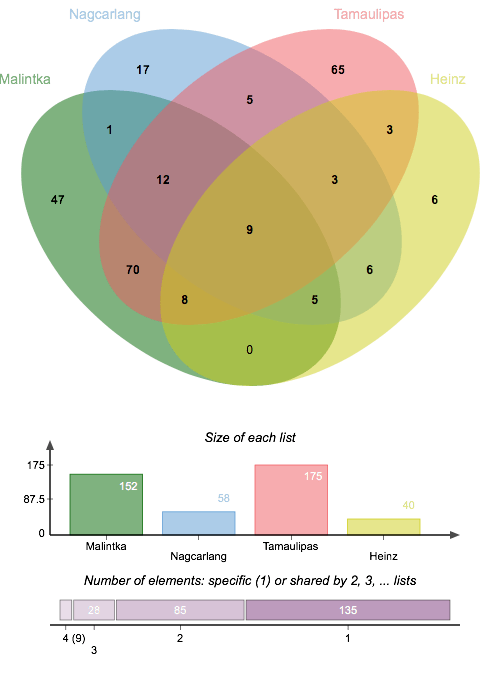
What do the patterns of differential gene expression between heat unstressed and stressed conditions for thermosensitive and thermotolerant cultivars of Solanum lycopersicum reveal about their heat stress responses? Using R code developed by thermotolerance Ann Loraine and with help from other collaborator, a list was generated of genes demonstrating significant differential expression between the control and heat stress groups for each cultivar. There is a high degree of overlap between the Malintka and Tamaulipas cultivars and less overlap for the Nagcarlang group with either of the other thermotolerant cultivars. Transport regulation, protein refolding, and potentially transcription are aspects of Nagcarlang’s unique response to heat stress. “Unique” in this context does not necessarily refer to the molecular mechanisms as being chemically dissimilar, but rather, that the expressed loci employed to produce these responses are different from Malintka and Tamaulipas.