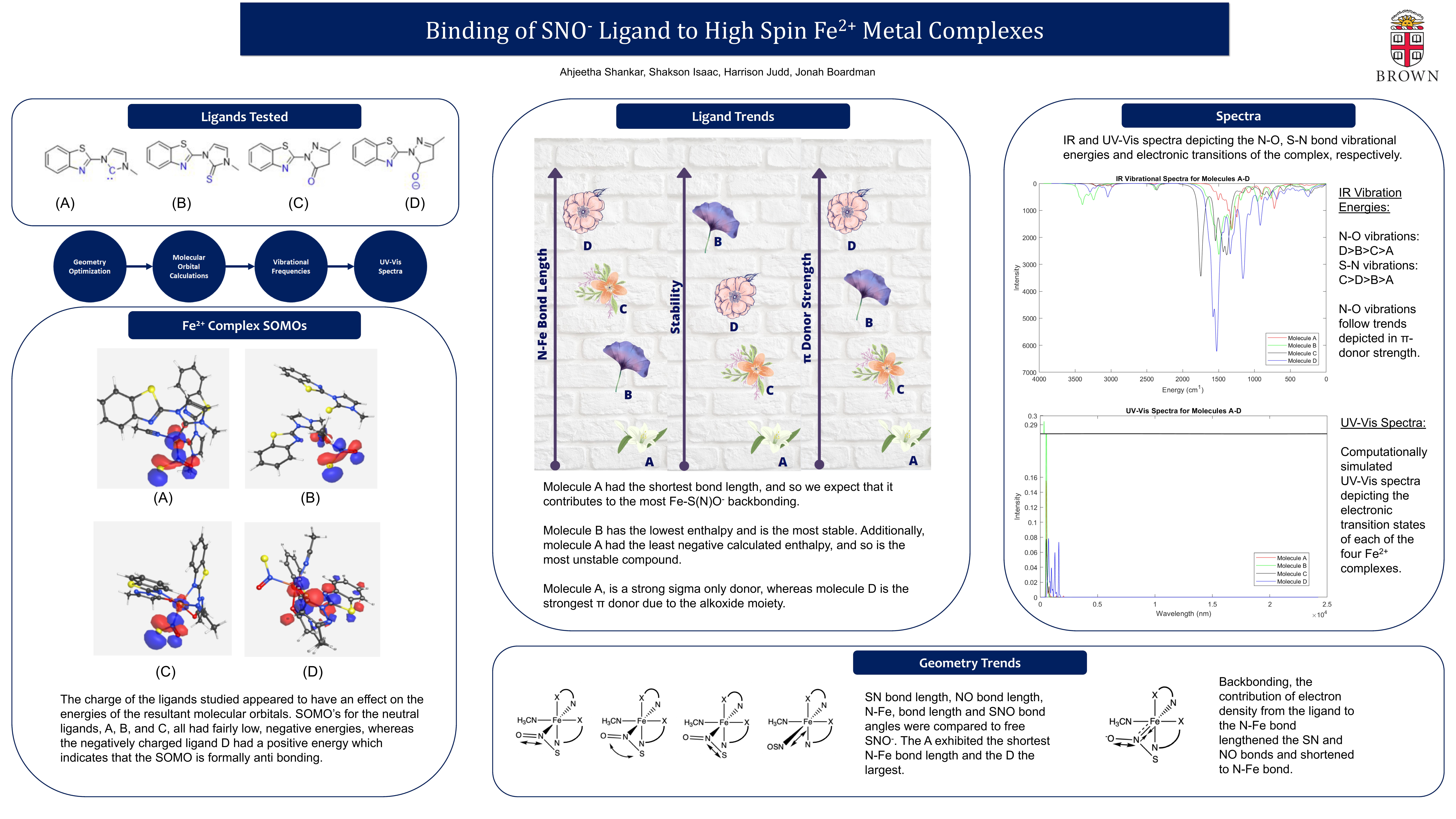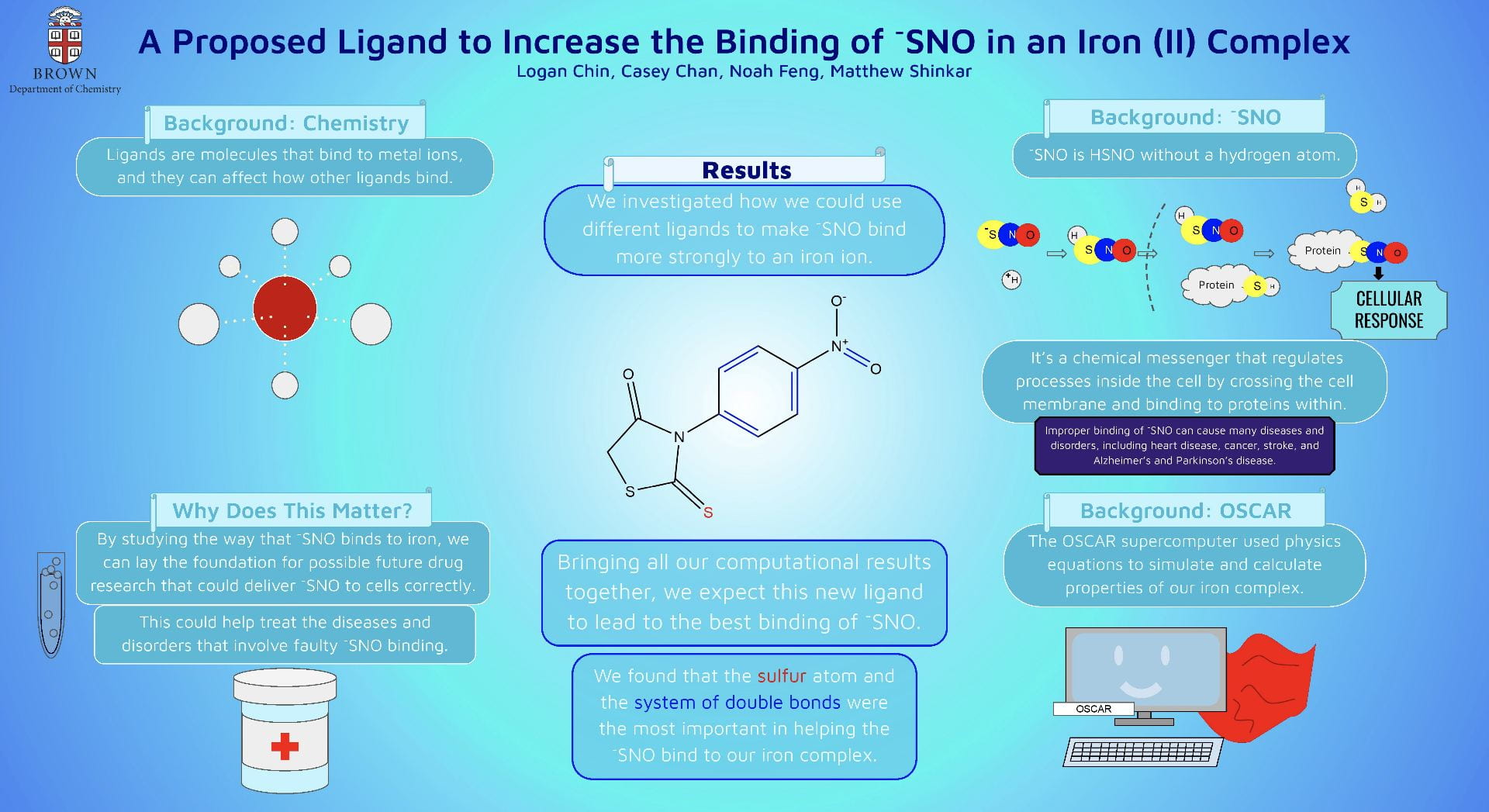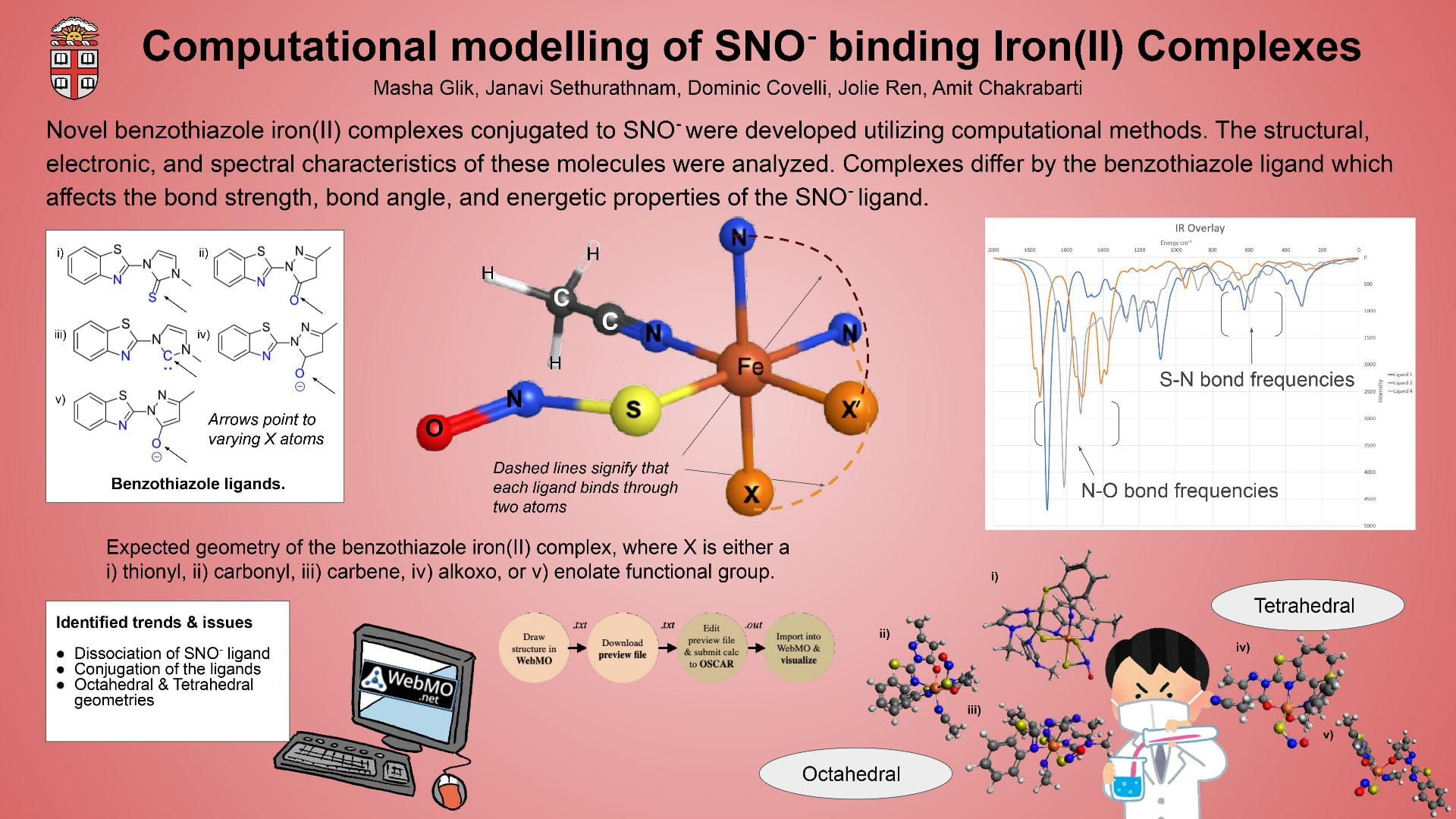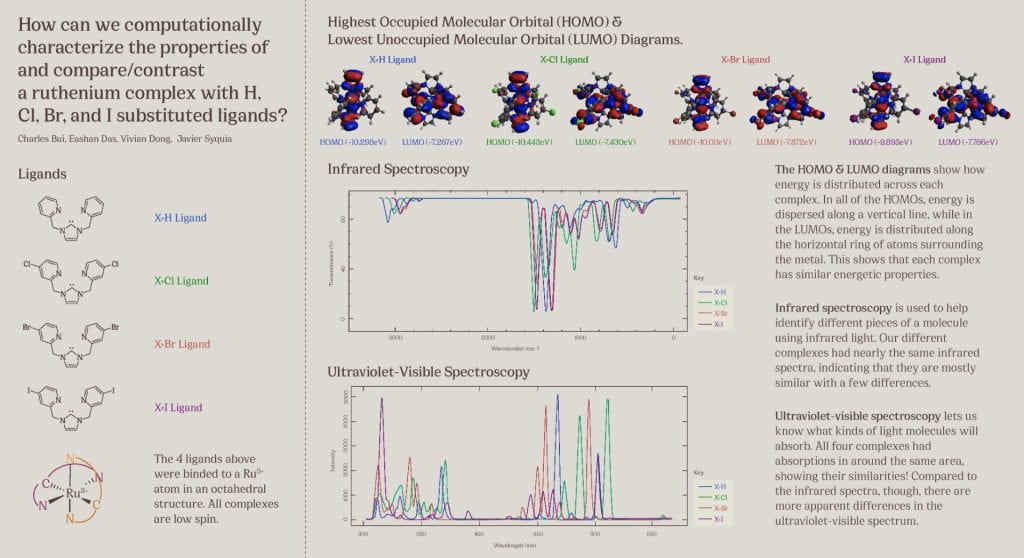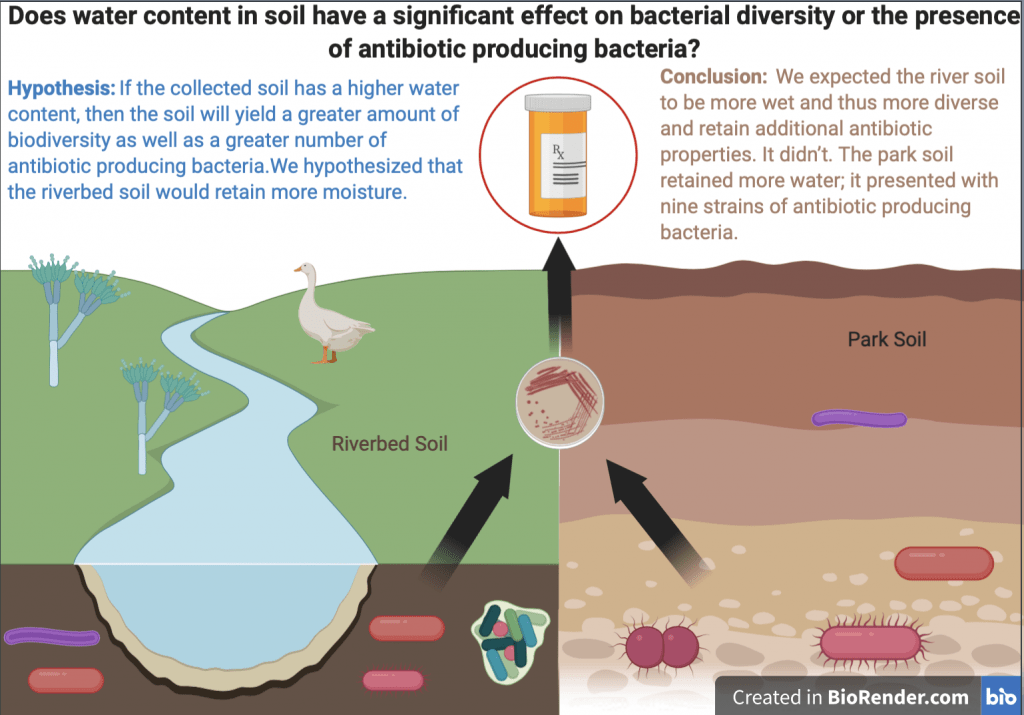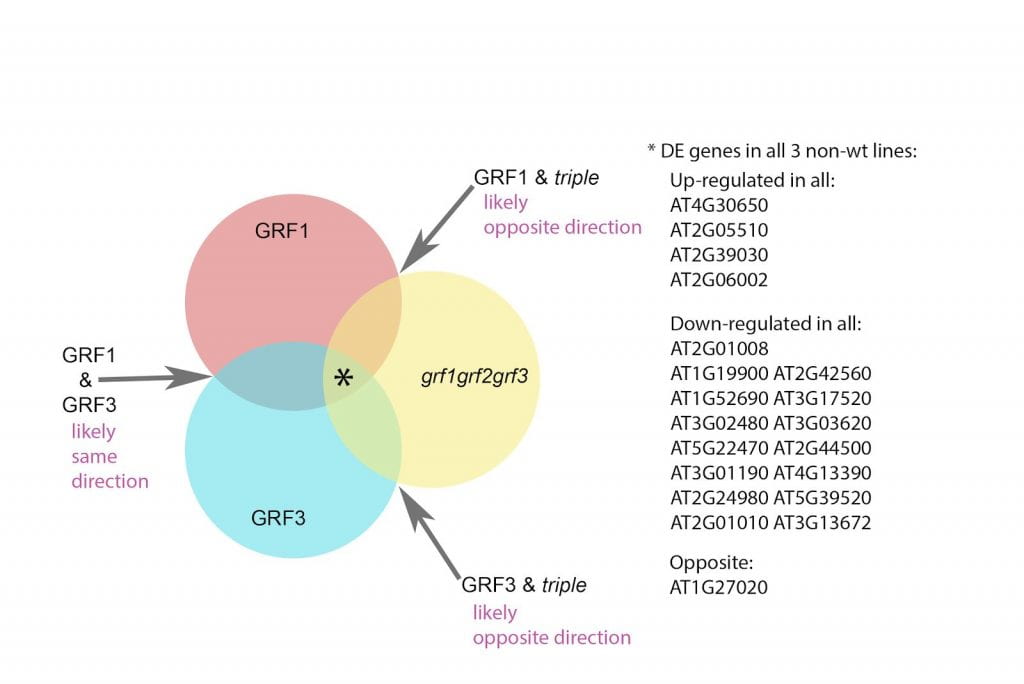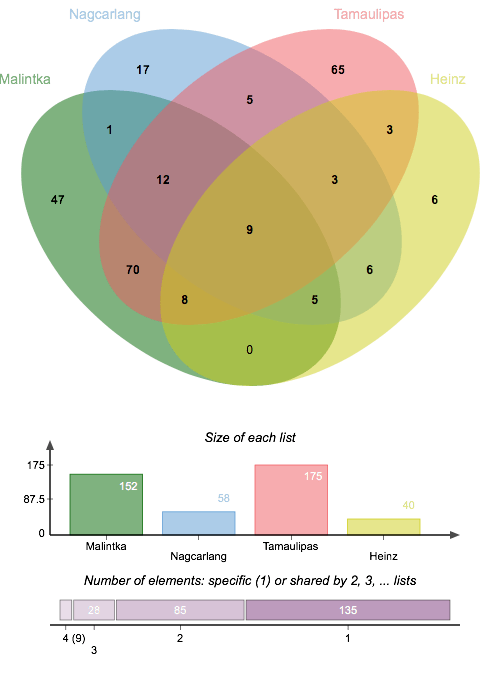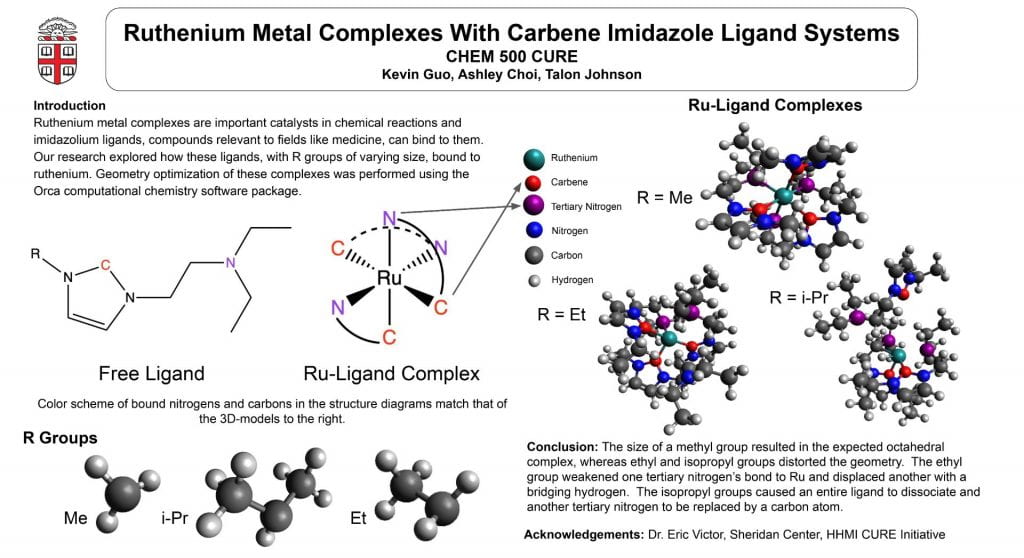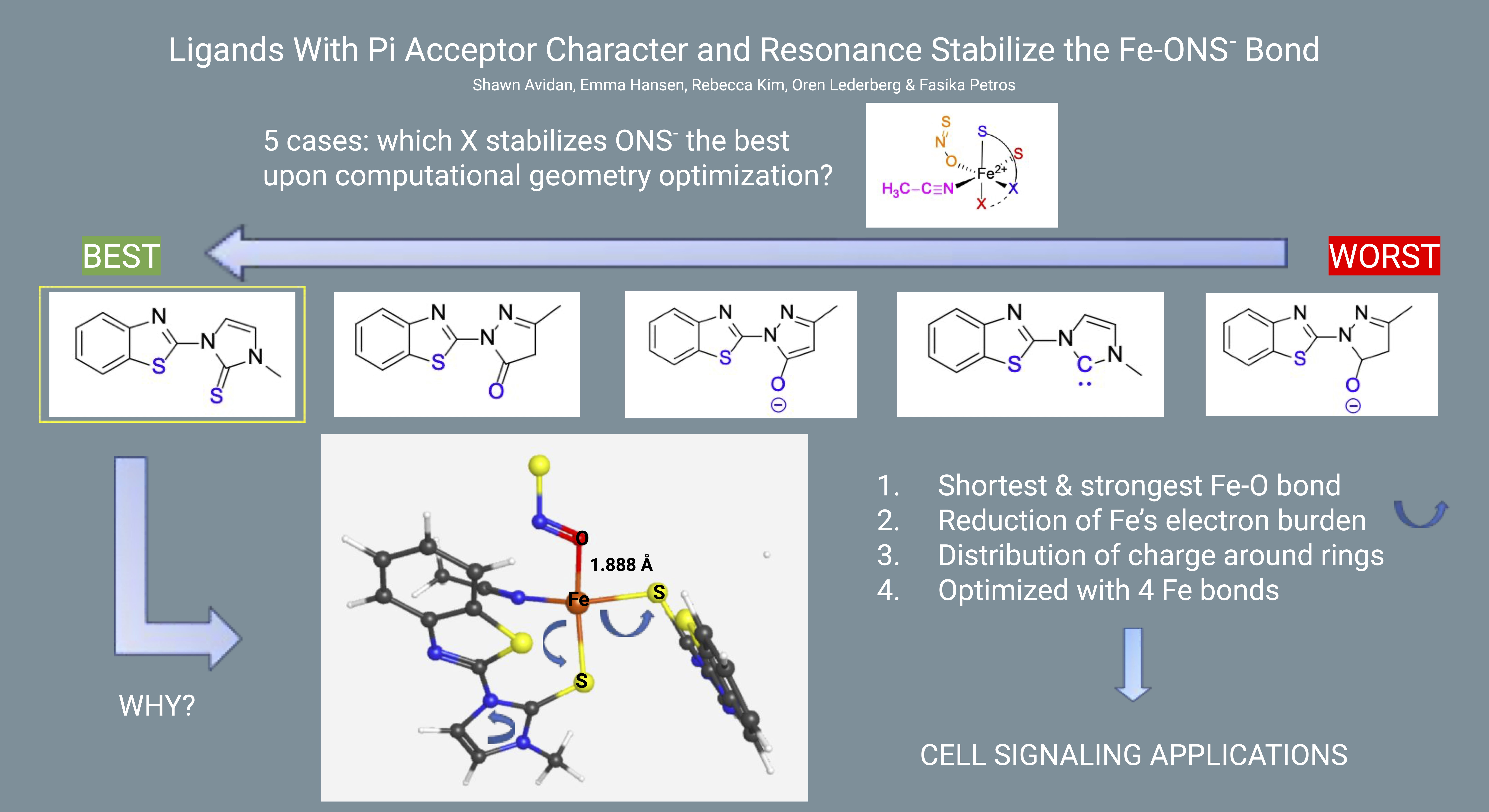
This poster displays the five ligands we examined in our experiment ranked in order of how well they stabilized an (SNO)- ligand, and a summary of our main findings.
Class: CHEM 0500 – Inorganic Chemistry Lab
Instructor(s): Dr. Eric Victor
Student(s): Rebecca Kim ’23, Emma Hansen, Fasika Petros, Oren Lederberg, Shawn Avidan (Chemistry ’22)
Description:
Our objective was to find a ligand that best stabilized an (SNO)- ligand coordinated to an iron center upon computational modeling. Computational optimizations included those for geometry, vibrational spectroscopy, UV-Visible spectroscopy, and molecular orbitals.
Using Fe-O bond length as a proxy to track stability, we discovered that a ligand with a 1) high degree of aromaticity and 2) pi-accepting character was best for stabilizing (SNO)-.
“CURE has increased my confidence about participating in research at Brown.”
-Student
I hadn’t really heard about CURE until taking this class. I appreciate the way CURE challenged me and I think CURE has increased my confidence about participating in research at Brown. It was a different experience from the typical lab portions of the class. I think there are advantages and disadvantages, it was unique and interesting and taught team building.