Class: BIOL 0440 – Inquiry in Plant Biology: Analysis of Plant Growth, Reproduction and Adaptive Responses
Instructor(s): Dr. Alison DeLong, Dr. Mark A Johnson
Student(s): Galen Tiong (Biology ScB ’20), Henry Dawson, Nicholas Moreno, Leane Pajot
Description:
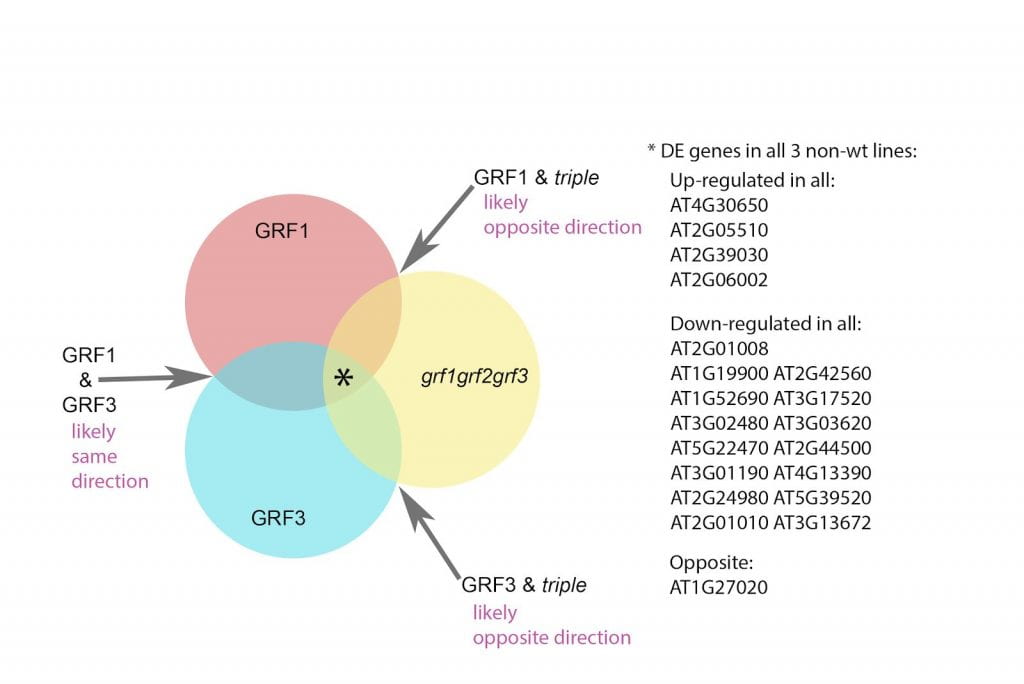
Growth regulating factors (GRFs) are known to regulate leaf size in plants in general, but little is known how different GRFs work in tandem with one another. Using bioinformatic tools, we analyze RNAseq data collected from different GRF mutants and examine which genes are differentially expressed (DEGs) as well as the overlap in regulation of gene expression among different mutants. We found a sizable DEG overlap between GRF mutants suggesting that GRF1 and GRF3 regulate expression in the same direction, whereas GRF2 might work as a negative regulator of GRF1. Our analyses also highlighted several genes of interest (e.g. PEPC1, GASA14) that could be direct targets of growth regulation and potential leads for future research into GRF function.